rna-tools.online
Minimize with QRNAS
files from another job e.g., 614e0a1b
The raw output files for each step of the pipeline can be found here and downloaded as a ZIP file here.
Documentation
rna_refinement - RNA refinement with QRNAS.
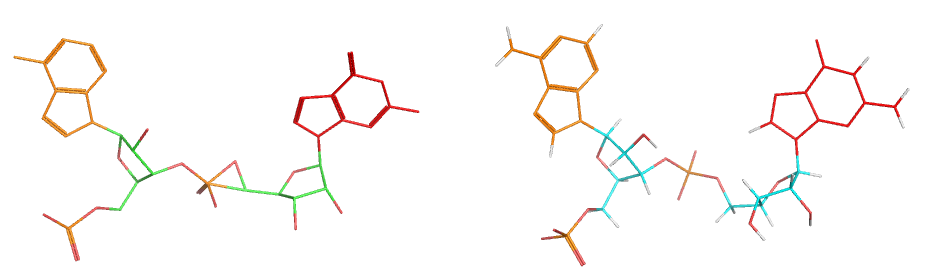
The demo input file with rebuilt phosphate is shown on the left, and the refined version ("_refx.pdb") on the left with a much better geometry of bonds.
Models of RNA 3D structures obtained by modeling methods often suffer from local inaccuracies such as clashes or physically improbable bond lengths, backbone conformations, or sugar puckers. To ensure high quality of models, a procedure of re nement should be applied as a nal step in the modeling pipeline. The software tool QRNAS was developed in our laboratory to perform local re nement of nucleic acid structures based on an extended version of the AMBER force field. The extensions consist of energy terms associated with introduction of explicit hydrogen bonds, idealization of base pair planarity and regularization of backbone conformation.
Read more: Piatkowski, P., Kasprzak, J. M., Kumar, D., Magnus, M., Chojnowski, G., & Bujnicki, J. M. (2016). RNA 3D Structure Modeling by Combination of Template-Based Method ModeRNA, Template-Free Folding with SimRNA, and Refinement with QRNAS. Methods in Molecular Biology (Clifton, N.J.), 1490(Suppl), 217-235. http://doi.org/10.1007/978-1-4939-6433-8_14J. Stasiewicz, S. Mukherjee, C. Nithin, and J. M. Bujnicki, “QRNAS: software tool for refinement of nucleic acid structures.,” BMC Struct. Biol., vol. 19, no. 1, p. 5, Mar. 2019.
We are exploring using https://piwik.pro to track usage of the server for grant proposals, scientific presentations. Please write to us if there is any problem with your job mail us [behind Apple Hide My Mail].
